About
The Bioinformatics Lab at University of Sannio is led by Luigi Cerulo and Francesco Napolitano, focusing on Systems Biology, AI-driven omics data analysis, and Computational Drug Discovery.
Systems Biology
Exploring biological data to uncover complex molecular mechanisms

AI-driven omics data analysis
Leveraging Artificial Intelligence to make biological predictions from omics data.

Computational Drug Discovery
Indentifying small molecules for clinical and laboratory applications.
Bioinformatics research activities at the University of Sannio were initiated in 2000 by Michele Ceccarelli and focused on the study of informatic processes in biological systems. As biological data began to grow exponentially, the lab focused toward data science approaches in bioinformatics under the co-direction of Luigi Cerulo from 2009. Starting in 2021, the Bioinformatics Lab has been led by Luigi Cerulo and Francesco Napolitano and employs advanced techniques to explore biological data, predict molecular mechanisms, and identify novel drugs and therapeutic targets, driving innovation in bioinformatics research.
News and Publications
06 May 2025
New Master Degree in Health Biotechnologies / Bioinformatics
A specialization in Bioinformatics is just starting within the framework of the new Master Degree in Health Biotechnologies, starting in Academic Year 2025/26!

17 Mar 2025
Prof. Marieke Kuijjer is visiting us!
Marieke Kuijjer, Associate Professor in Cancer Data Science, iCAN Flagship, University of Helsinki, Finland, and Group Leader of the Computational Biology and Systems Medicine group at the Centre for Molecular Biosciences and Medicine Norway (NCMBM), Nordic EMBL Partnership, University of Oslo, Norway, visited our Lab for scientific exchanges. She presented her work on gene networks rewiring in cancer during a seminar to the Department of Science and Technology. This was a very productive exchange!

19 Dec 2024
BioinfoLab on UniSannio Magazine
What is Bioinformatics and what do we do at UniSannio BioInfolab? We were asked these questions by UniSannio Magazine and were glad to answer!

16 Dec 2024
Welcome Martina!
Martina Palummo just joined our team as a postdoctoral researcher! More about her in the Team section below.

20 Nov 2024
Futuro Remoto
The lab will be present at FuturoRemoto 2024 to showcase bioinformatics applications to middle and high-school students.

15 Nov 2024
Welcome Mirella!
Mirella Sangiovanni just joined our team as a visiting researcher! More about her in the Team section below.

20 Oct 2024
Welcome Erica!
Erica Cogliano just joined our team as an intern! More about her in the Team section below.

15 Sep 2024
Welcome Marco!
Marco Nigro just joined our team as a postdoctoral researcher! More about him in the Team section below.
10 Sep 2024
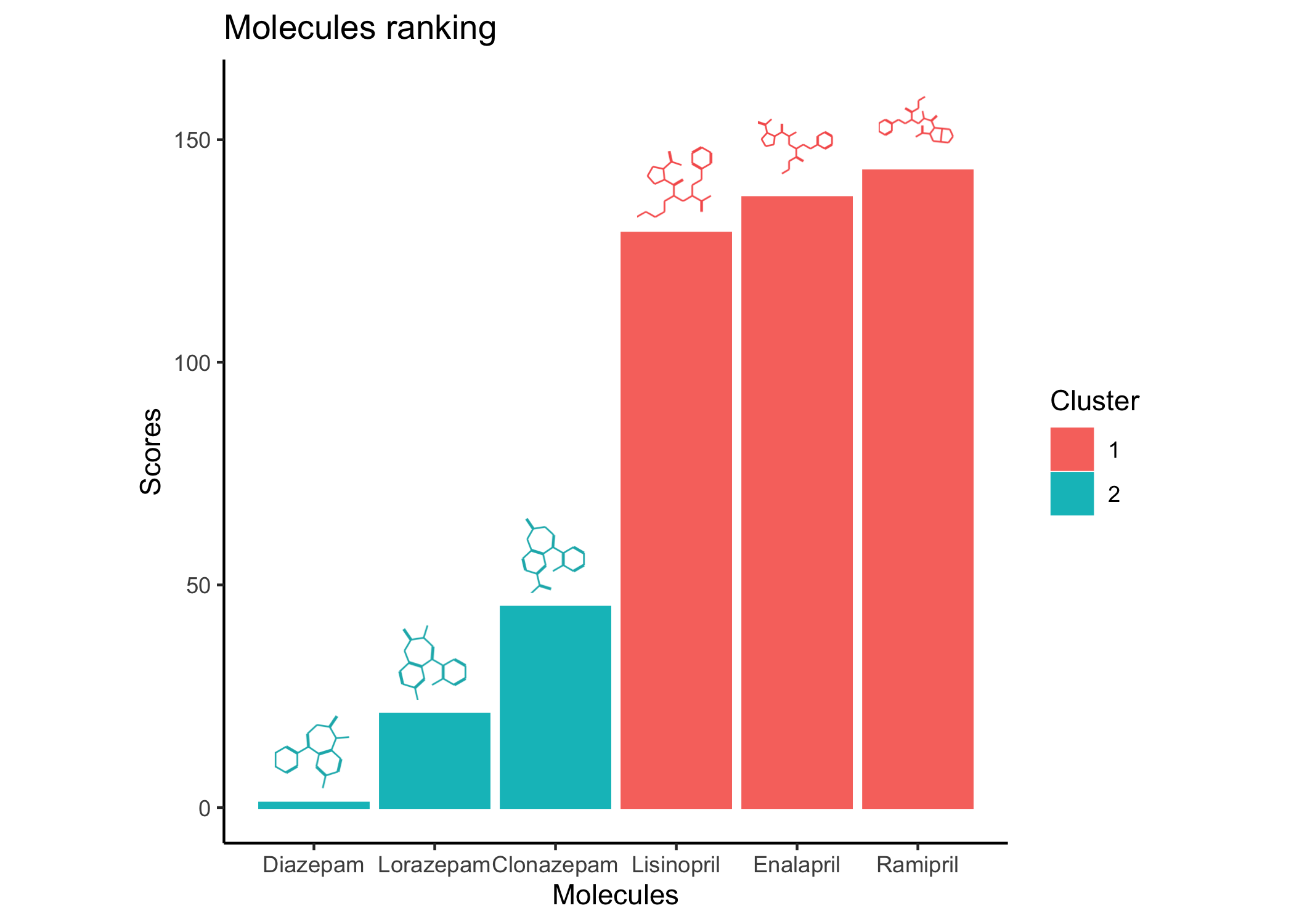
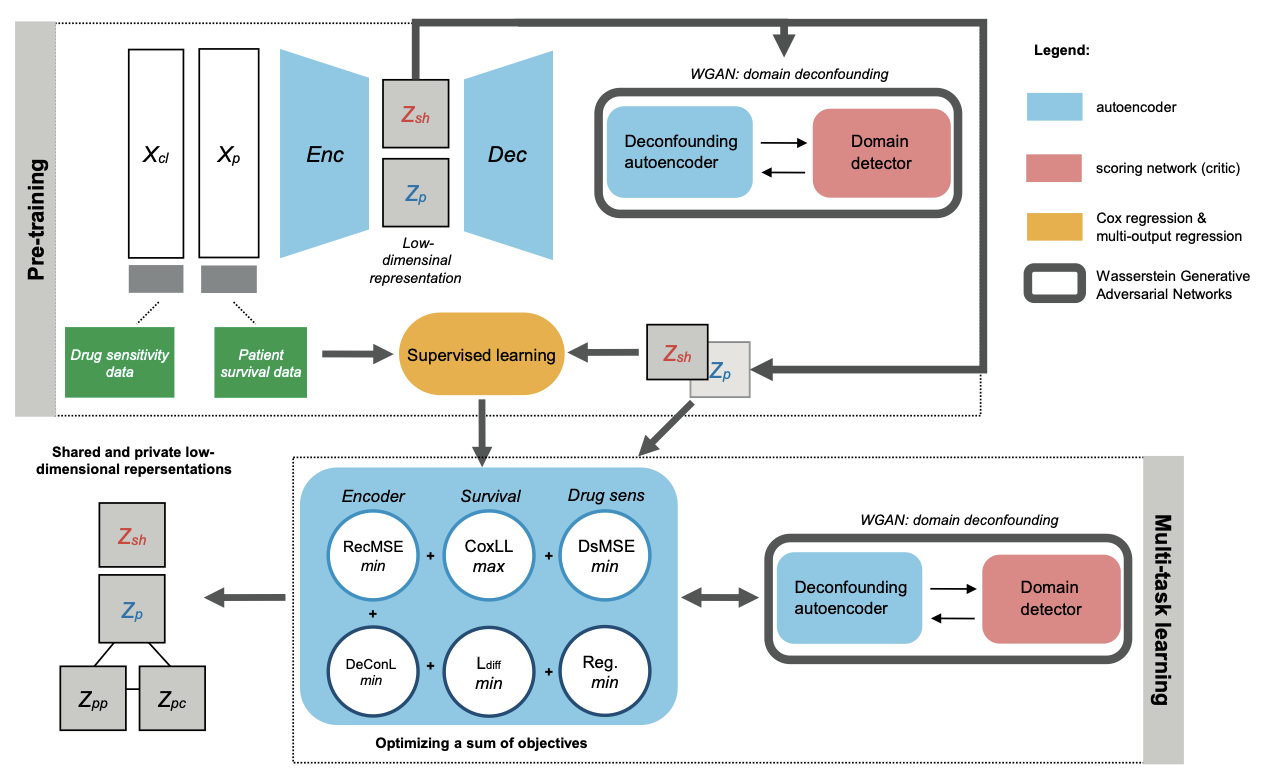
"Multi-task deep latent spaces for cancer survival and drug sensitivity prediction", in Bioinformatics

09 Sep 2024
CIBB 2024 was great!
The 19th conference on Computational Intelligence methods for Bioinformatics and Biostatistics has just finished and was a great success! Find more info (and pictures) at http://cibb2024.unisannio.it!
29 Jul 2024
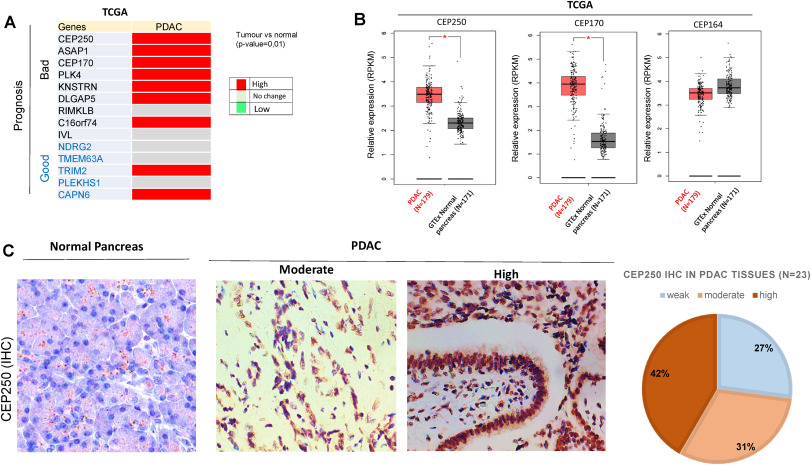
"Altered centriolar cohesion by CEP250 and appendages impact outcome of patients with pancreatic cancer", in Pancreatology
01 May 2024
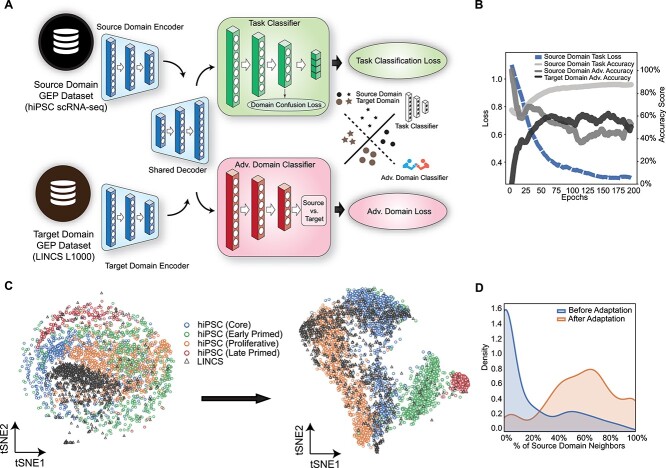
"AI identifies potent inducers of breast cancer stem cell differentiation based on adversarial learning from gene expression data", in Briefings in Bioinformatics
15 Mar 2024
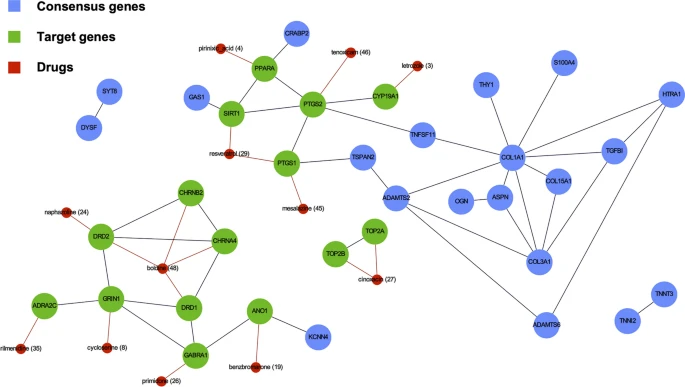
"Identification of therapeutic targets in osteoarthritis by combining heterogeneous transcriptional datasets, drug-induced expression profiles, and known drug-target interactions expression data", in Journal of Translational Medicine

01 Feb 2024
We are hosting CIBB 2024!
We are organizing the 19th conference on Computational Intelligence methods for Bioinformatics and Biostatistics. Find more info at http://cibb2024.unisannio.it!
Tools
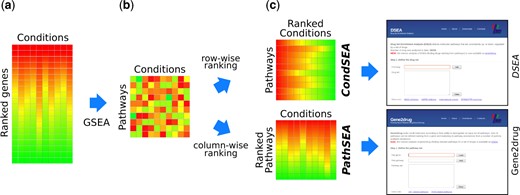
Gep2pep R/Bioconductor package
Pathway Expression Profiles (PEPs) are based on the expression of pathways (defined as sets of genes) as opposed to individual genes. This package converts gene expression profiles to PEPs and performs enrichment analysis of both pathways and experimental conditions, such as “drug set enrichment analysis” and “gene2drug” drug discovery analysis respectively.
Facilities
-

Informatics lab
50 desktop computers equipped with LTS Ubuntu and major bioinformatics tools for laboratory activities.
-

Superdome Flex
224 CPU cores Intel(R) Xeon(R) Platinum 8280 CPU @ 2.70GHz, 1.5 TB RAM, 150TB storage, 1 GPU NVIDIA Tesla V100.
-

Server cluster
Cluster of 7 CPU servers and 1 GPU unit including a total of 400 CPU cores, 4 TB RAM, 600TB storage, 6 GPU NVIDIA. Hosted at Biogem - Molecular Biology and Genetics Research Institute in Ariano Irpino (AV).
People
Current members

Luigi Cerulo
Lab Director

Francesco Napolitano
Lab Director

Marco Nigro
Postdoctoral Fellow

Antonio Ammendola
PhD student

Luca Faretra
PhD Student

Martina Palummo
Postgraduate Fellow

Diana Parente
Intern

Mirella Sangiovanni
Visiting PhD student

Erica Cogliano
Intern

Chiara Circelli Gabriele
Bachelor Student

Manuel Maio
Bachelor Student
Former members

Donatella Pierri
Bachelor student

Francesco Saccomando Ciaramella
Intern, Bachelor student

Felicita Masone
Bachelor student